It is a very good article to help you better understand the reference genome that we are using daily...l
What is a genome assembly?
The haploid human genome consists of 22 autosomal chromosomes and the Y and the X chromosomes. Each of the chromosomes represents a single DNA molecule, a sequence of millions of nucleotide bases. These molecules are linear, so one might expect that we should represent each chromosome by a single, continuous sequence. Unfortunately, this is not the case for two main reasons: 1) because of the nature of genomic DNA and the limitations of our sequencing methods, some parts of the genome remain unsequenced, and 2) emerging evidence suggests that some regions of the genome vary so much between individual people that they cannot be represented as a single sequence. In response to this, modern genomic data sets present a model of the genome known as a genome assembly. This post will introduce the basic concepts of how we produce such assemblies as well as some basic vocabulary.
One of the major challenges in sequencing eukaryotic chromosomes is their sheer size, making it impossible with current technology to obtain their sequences by progressing from one end to the other. Instead, researchers commonly isolate genomic DNA from a biological sample and fragment the DNA into small pieces that can be sequenced individually. These individual, contiguous sequences are called reads, and generally number from 100 to 1000 nucleotide bases depending on the technology. Researchers then assemble the reads, like pieces of a giant puzzle, into progressively larger contiguous pieces. Finally, they assemble these larger pieces into full chromosome sequences.
In the Human Genome Project (HGP), researchers used a variation of the general method above called a clone-based (or hierarchical) approach (see below for links to additional information). This method used a library of fairly large, overlapping segments of genomic DNA. These segments are called clones and generally contained 40,000 – 200,000 nucleotide bases. Researchers then inserted these clones into vectors, such as bacterial artificial chromosomes (BACs), and propagated them in bacteria to generate large amounts of DNA for sequencing. They then sequenced the individual clones by fragmenting them into reads, sequencing the reads and then assembling the reads to produce the full clone sequences, which are called components. They used additional mapping methods to determine which of the components overlapped, and then used these data to organize the components into a linear arrangement known as a tiling path. HGP researchers continued assembling the sequence into larger units as long as there were overlaps between individual components. The resulting “top-level” sequences that have no gaps and represent a continuous region of a chromosome are called contigs orscaffolds. To generate chromosome models, HGP researchers ordered and joined the scaffolds that belonged to individual chromosomes.
An example: human chromosome 17
Let’s take a look at an example. Figure 1 shows a portion of the tiling path of human chromosome 17 (GenBank record CM000679.1). This region of the chromosome was based on six components, each of which has its own accession number at NCBI (e.g.AC006236.2), and these records contain the complete sequence data for each component. In this region, the tiling path is an unbroken series of overlapping components.
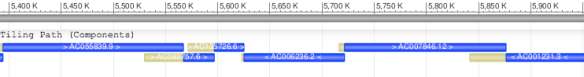
Figure 1: Graphic representation of small portion of the tiling path of human chromosome 17. The assembled chromosome is shown in grey, the individual components in blue, and the areas of overlap in yellow.
Let’s look at another portion of chromosome 17 (Figure 2). This region is near the centromere of the chromosome, and you’ll see that the tiling path is broken near position 21,570 K. With the gaps present, you might wonder why the chromosome sequence (the grey bar) is unbroken. In fact, if you look in the actual chromosome record (CM000679.1), you’ll only find a series of “N”s in that gapped segment instead of the normal bases (A, C, T or G). Representing gaps with “N”s allows us to generate a single, continuous record for an incompletely assembled chromosome while also clearly showing where the gaps in the data lie.
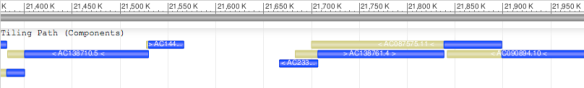
Figure 2: Graphic representation of the tiling path near the centromere of human chromosome 17. The assembled chromosome is shown in grey, the individual components in blue, and the areas of overlap in yellow.
Why are there still gaps in the assembly?
Since the initial release of the human reference genome in 2001, researchers have made great strides in improving the quality of the assembly model, but significant challenges remain. One of these is the simple fact that certain regions of genomic DNA are much more difficult to sequence than others. For example, the telomeres and centromeres of chromosomes contain tightly packed DNA known as heterochromatin, and these regions are difficult to sequence because of the high frequency of G and C bases. Genomic DNA also contains many repetitive sequences, and this makes assembling sequencing reads a daunting task. Imagine taking the text of the beloved children’s book “Green Eggs and Ham” (with many repetitive words), shredding it and then trying to assemble the sentences back in the right order. There are many possible solutions that will generate grammatically correct English sentences, but none of these (save one) will be the correct Dr. Seuss original. Another complicating factor is that the HGP DNA samples came from multiple people, so that the resulting “genome” is really a randomly mixed conglomerate that, in some cases, may be impossible to represent correctly as a single sequence. We are now much more aware that some regions of the genome can vary quite dramatically from individual to individual, and this new awareness is helping to guide new genome assemblies. Since 2007, these improved assemblies are the responsibility of the Genome Reference Consortium (GRC). In future posts, we will have a look at the GRC’s effort to improve and modernize the assembly, and discuss the relevance of a high-quality reference assembly for the NGS sequencing projects.
Citations and literature guide
History of the Human Genome Project: key news releases and publications
- February 12, 2001: International Human Genome Sequencing Consortium Publishes Sequence and Analysis of the Human Genome
- Lander, E.S. et al. (2001) Initial sequencing and analysis of the human genome.Nature, 409, 860-921.
- April 14, 2003: International Consortium Completes Human Genome Project
- October 20, 2004: International Human Genome Sequencing Consortium Describes Finished Human Genome Sequence
- International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature, 431, 931-45.
Glossary terms
The links below lead to alphabetically indexed pages within the Gene Reviews Illustrated Glossary or the Genetics for Surgeons Glossary. Please scroll to the term on the page.

Genome sequence assembly is a core problem in bioinformatics. The sequence generates sequencing reads with sequence assembly, and the base sequence of the genome is generated.
ReplyDeleteRelated link: http://www.cd-genomics.com/Whole-Genome-SNP-Genotyping-Service.html