I was aware of several software can be used to deal with this type of data:
Tao Liu's MACS https://github.com/taoliu/MACS/
this is probably the most widely used method for ChIP-seq peak calling, and it is also suitable for ChIP-exo data
https://github.com/taoliu/MACS/issues/15
Peakzilla https://github.com/steinmann/peakzilla
A new tool for ChIP-exo
"Peakzilla identifies sites of enrichment and transcription factor binding sites from transcription factor ChIP-seq and ChIP-exo experiments at hight accuracy and resolution. It is designed to perform equally well for data from any species. All necessary parameters are estimated from the data. Peakzilla is suitable for both single and paired end data from any sequencing platform."
A quick google I found several others:
MACE http://chipexo.sourceforge.net/
and GEM http://www.psrg.csail.mit.edu/gem/
Since MACS (version 1.4) was pre-installed in the HPC at UFL, I decided to give it a try
[mtang@dev1 mm10]$ module load macs
macs -t ChIP.bam -c control.bam -f BAM -g mm -n ChIP-exo -B
it took some time to finish the process (~30mins). with -B flag, I want to output the bedgraph files for each chromosomes. If you specify -S, it will give you a single bedgraph file for the whole genome.
Look at the model built by MACS
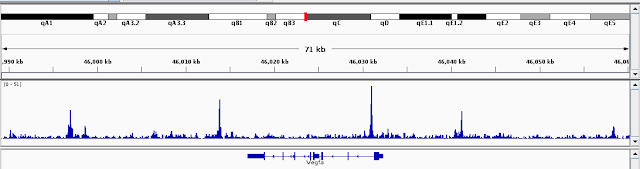
You can have a quick look at the data by loading the bedgraph files into IGV. I just pick VEGFa to check
The peaks look very specific and sharp.
Then, I can find all the genes that contain a peak nearby.
Homer annotatePeaks http://biowhat.ucsd.edu/homer/ngs/annotation.html
if you use R, ChIPpeakAnno http://www.ncbi.nlm.nih.gov/pubmed/20459804
cistrome can do it very easily http://cistrome.org
BETA-minus: Targets prediction with binding only Predict the factors (TFs or CRs) direct target genes by only binding data
CEAS http://liulab.dfci.harvard.edu/CEAS/ also from Liu's lab
the easiest way PAVIS http://manticore.niehs.nih.gov:8080/pavis/


This comment has been removed by a blog administrator.
ReplyDelete